
In drug discovery, choosing the wrong target is often the most expensive mistake a company can make. It can mean years of effort, millions of dollars, and ultimately failed clinical trials. The root cause? Many traditional discovery methods simply miss the genes that really matter.
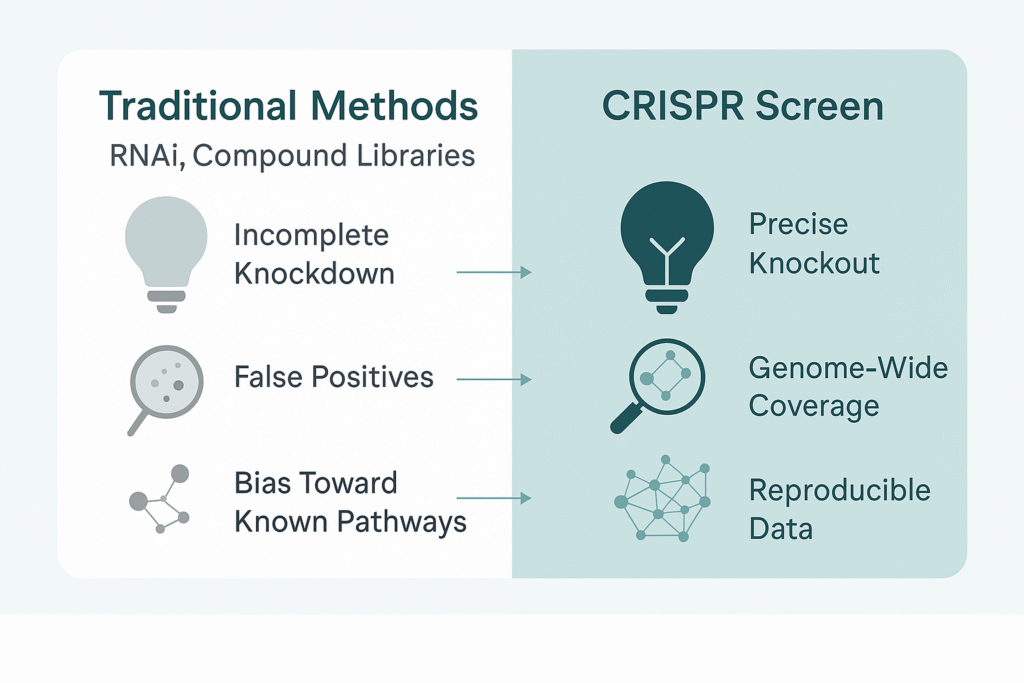
For years, RNAi and compound library screening were the standard tools for functional genomics. They played their part, but both suffer from critical flaws: incomplete knockdowns, high false positives, and a bias toward known pathways. These blind spots mean drug developers may walk straight past the very targets that could unlock a breakthrough.
CRISPR screening is changing that. With higher specificity and genome-wide coverage, CRISPR screens can systematically reveal the functional drivers of disease—drivers that traditional methods routinely overlook. The ability to discover, prioritize, and validate hidden targets is giving research teams and pharma companies a smarter way forward.
Why Traditional Methods Miss Key Targets
The limits of RNAi/shRNA
RNAi once revolutionized target discovery, but its flaws became clear. Knockdown efficiency is inconsistent—some genes are only partially silenced, producing weak phenotypes and false negatives. Off-target activity, on the other hand, triggers misleading results and false positives. In practice, teams spend months chasing noise instead of true hits.
The limits of compound libraries
Compound libraries rely on known molecules and known pathways. They may show an effect, but rarely reveal whether the compound touches the root cause of disease. For exploratory projects, or when seeking novel targets, these libraries are blunt instruments.
Impact on drug developers
The consequences are severe:
- High attrition: Over 80% of oncology projects fail before Phase II, often due to poor target validation.
- Runaway costs: Every wrong lead adds months and millions.
- Lost opportunity: Competitors who identify true drivers first capture the market advantage.
The industry doesn’t just need new targets—it needs more reliable ways to find them.
What Is a CRISPR Screen and Why It’s Different
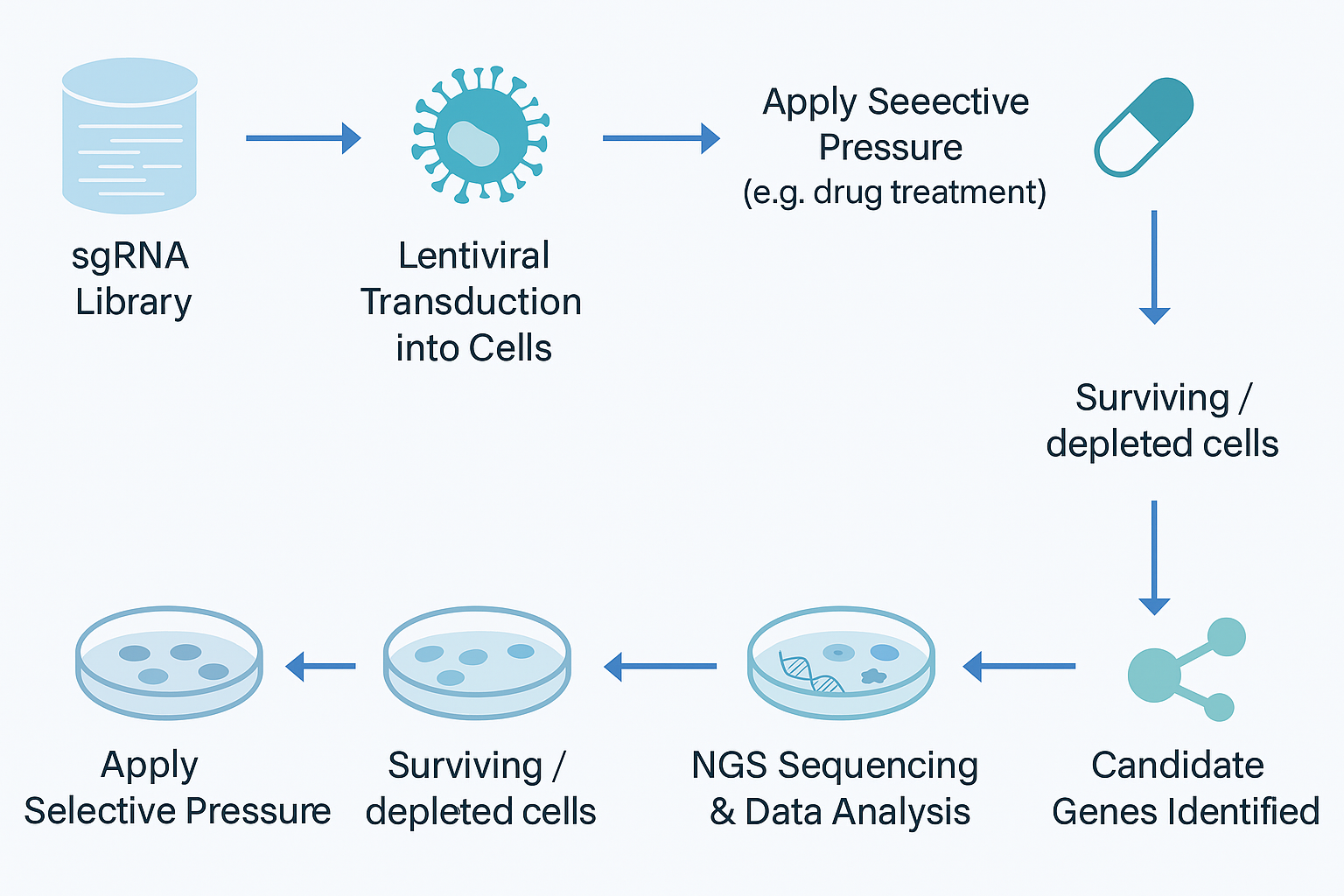
At its core, a CRISPR screen is a large-scale experiment where thousands of guide RNAs are used to knock out, repress, or activate genes across the genome. By applying a selective pressure—drug exposure, viral infection, or stress—and comparing outcomes, researchers can identify the genes that truly matter.
If you want a practical overview of how a CRISPR screen works in research, click here.
Here’s why this approach is different from older tools:
- Higher specificity: CRISPR knockout is closer to “lights fully off,” while RNAi is often “half-dimmed.”
- Broader coverage: Entire genomes can be screened, or sub-libraries can focus on specific pathways.
- Flexible strategies: CRISPRi suppresses expression without DNA breaks, while CRISPRa activates genes. This extends discovery to non-coding and regulatory regions.
- Greater reproducibility: Lower noise means higher validation rates—critical for pharma investments.
Think of CRISPR screening as moving from a fuzzy map to a high-resolution GPS. Suddenly, the landmarks are clear.
How CRISPR Screening Reveals Hidden Drug Targets
Hidden targets often hold the most value. CRISPR screening reveals them across multiple domains:
Oncology: separating drivers from passengers
Tumors are riddled with mutations, but only a fraction drive malignancy. CRISPR screens distinguish the true drivers. For example, genome-wide screens have identified novel oncogenes like NCAPG, which traditional tools missed but later proved essential for tumor growth.
Drug resistance mechanisms
Every targeted therapy faces resistance. Traditional methods discover resistance only after it arises. CRISPR screens predict resistance genes up front—for example, uncovering NF1 and CUL3 in melanoma resistance to vemurafenib. Knowing these risks early helps teams design combination therapies that last.
Immunotherapy optimization
Responses to checkpoint inhibitors vary dramatically. CRISPR screens reveal the genes shaping T cell activity or tumor microenvironment interactions. By exposing regulators of immune evasion, they point the way to more precise—and more predictable—therapies.
Infectious disease
Beyond oncology, CRISPR screens have mapped host factors for viruses like dengue, West Nile, and hepatitis C. Identifying host dependency genes enables development of broad-spectrum antivirals—targets that traditional compound libraries never touch.
Where older tools stumble in the dark, CRISPR screening turns on the light.
From Screening to Validation: The Role of CRISPR Gene Editing
Screens generate candidate lists. Validation separates discovery from wishful thinking. This is where CRISPR gene editing becomes indispensable.
- Knockout studies: Does deleting a candidate gene make cells more drug-sensitive?
- Knock-in or rescue experiments: Can reintroducing the gene reverse the phenotype?
- Point mutations: Does a single nucleotide change alter response, mirroring clinical variants?
Consider oncology again: a genome-wide CRISPR screen may yield hundreds of candidates. Systematic knockout validation can reduce that to a handful of credible drivers worth further investment.
For a deeper look at how CRISPR gene editing platforms accelerate validation and model building, click here.
Together, CRISPR screen plus CRISPR gene editing create a closed loop—discovery plus confirmation. For pharma teams, this loop means faster, more confident go/no-go decisions, saving years of wasted effort.
Designing a Reliable CRISPR Screen
A screen is only as strong as its design. Poor planning wastes time and money. Here are key principles:
- Define the objective clearly: Are you searching for essential genes, resistance mechanisms, or regulators of a pathway? Positive or negative screen? Clarity here shapes everything that follows.
- Choose the right library: Genome-wide for discovery, sub-libraries for hypotheses, custom designs for efficiency.
- Ensure sufficient coverage: Aim for 500–1000× coverage. A 20,000-sgRNA library means at least 10 million cells. Undercoverage causes dropout bias, where guides disappear not from biology but lack of representation.
- Optimize transduction and culture: Keep MOI near 0.3 so that most cells carry only one sgRNA. Stable culture conditions reduce noise.
- Analyze and validate rigorously: Tools like MAGeCK and BAGEL highlight enriched genes, but only functional validation turns lists into actionable evidence.
Example: One DNA repair screen produced 400 initial hits. After applying coverage filters and knockout validation, only 18 proved reproducible. That difference—between 400 and 18—is the line between noise and true discovery
Recommended Tools and Products
Running a CRISPR screen from scratch is challenging. Proven resources help remove bottlenecks:
- CRISPR Libraries (In-stock & Customized): Ready-to-use genome-wide or pathway-focused sets save months of setup.
- gRNA Plasmid Bank: Pre-built plasmids for rapid validation shorten timelines.
- KO Cell Line Bank: Thousands of knockout lines let researchers test phenotypes immediately.
- Cas9 Stable Cell Lines: Prebuilt Cas9 models cut prep time by 3–5 weeks, improving reproducibility.
- Optimized Media and Reagents: Specialized culture solutions maintain cell health and representation.
Each tool addresses a pain point, helping teams focus on science instead of logistics.
Conclusion: A Smarter Path to Drug Discovery
Drug discovery succeeds or fails on target choice. Traditional tools left blind spots that cost time, money, and competitive advantage. CRISPR screening changes that by illuminating the targets that really matter.
When paired with CRISPR gene editing, screening moves from discovery to validation, turning lists into evidence and evidence into confident decisions. With ready-made libraries, knockout lines, and stable Cas9 platforms, teams no longer need to reinvent the wheel—they can move straight to answers.
Adopting CRISPR screening isn’t about following a trend. It’s about avoiding the 80% attrition rate that haunts traditional pipelines. For research groups and pharma companies aiming to compete in today’s market, the question is simple: are you still relying on tools that miss half the story, or are you ready to illuminate the hidden targets that matter most?




